When you say restriction enzymes cut covalent bonds (within a single strand), do you mean that they cut the bonds that connect adjacent nucleotides of a double-stranded DNA molecule? Or what exactly do you mean by within a single strand?
They cut adjacent nucleotides within a strand, yeah. So:
ATAATATA|ATA
Cuts the AA bond - the hydrogen bond to the other strand is broken as a result of this.
Also, when trying to transport a DNA fragment into another cell, say a bacterial cell. Why do we only have to use restriction enzymes with sticky ends to cut the passenger-DNA fragment, and then use the same restriction enzyme to cut the plasmid? Is it because by using the same restriction enzyme, it will create the appropiate stick ends on the plasmid as there is on the passenger-DNA fragment, and in that case they can join together when mixed and under the presence of ligase?
I'm not really sure what the question is here - you might be getting mixed up with transformation (which isn't covered in VCE?)
But yeah, if you cut insert/fragment A and vector/plasmid A with restriction enzyme B, you will generate complimentary sticky ends which can then be ligated together. The complete ligated vector can then be transformed into competent cells.
When ligase joins together two fragments of DNA, does it only catalyse the binding of the phosphate group with the hydroxyl group, or does it also bind together the hydrogen bonds between the complementary bases?
It only catalyses the formation of the phosphodiester bond. The hydrogen bonds between the two sticky ends must already have formed. The formation of the hydrogen bonds can happen without an enzyme.
(Ligase reactions are generally run at a lower temperature to facilitate this.)
Also also, why can't ligase join together two blunt ends? I understand that they don't have the bases sticking out, and so how can the two pieces of DNA join, well can't ligase actually bind the phosphate group of one DNA fragment, directly to the sugar of the other one, without there being any base-pairing?
You can do 'blunt ended ligations,' but they are very difficult, as you might imagine. You do these reactions in conditions to facilitate an interaction between the two blunt ended strands. These reactions are difficult to conduct, and you end up with a bunch of products.
Ligating sticky ends is much easier and you should only end up with one product (the plasmid you're interested in.)
Thank you Maca 13
1). When an enzyme's disulphide bridges are broken, what will happen to the enzyme?
I said the shape of the active site will obviously by altered, and hence the enzyme can no longer react and bind to it's specific substrate as it is no longer complementary.
However, should the word 'denatured' be included? Has the enzyme been denatured or not? I know if the enzyme was exposed to factors outside it's optimum range it would become denatured (besides low temperatures), so does that mean enzyme's become denatured when their overall shape is changed, and hence the bonds in the active site is are broken?
Denature would be fair. I suspect that the answer is actually more complex, because of the role that disulphide bonds actually play in protein folding, but thinking with my VCE hat on, I daresay denaturation would be a fair assumption. Maybe if mahler's around he'll shed some light on this 
Most of the time, it's probably fair to say that if you remove a disulphide bond, a protein's function will be altered. There's also a good chance that the protein will not fold correctly, and may be insoluble.
Be careful with conflating a 'change of shape' with denaturation. Enzymes/proteins often change shape ('conformation'). These conformational changes are very important in protein function - an example you may have come across is haemoglobin - oxygen-bound haemoglobin has a different conformation to deoxyhaemoglobin. The oxygen-bound confirmation adopts a confirmation which has a higher affinity for oxygen. Both deoxy and oxy haemoglobin do have structure however, it's just that the shape changes through the course of the protein's function. Indeed, there are now a lot of examples of proteins that don't have structure - that are still functional!
When you denature an enzyme, you lose all structure - the protein adopts a 'random coil' structure which is not biologically active:
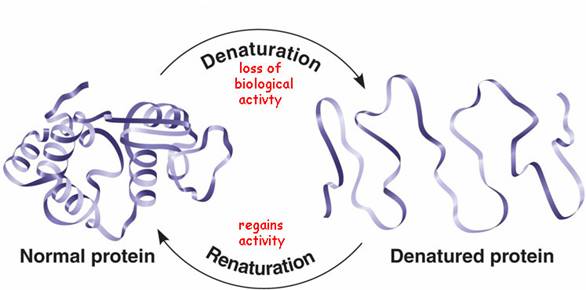
As this picture suggests, this process is often (but by no means always) reversible.
[Obligatory 'more detail then you need for VCE, but useful learning for life.']

